Page 14 - BMC_1471
P. 14
BMC Evolutionary Biology 2008, 8:56 http://www.biomedcentral.com/1471-2148/8/56
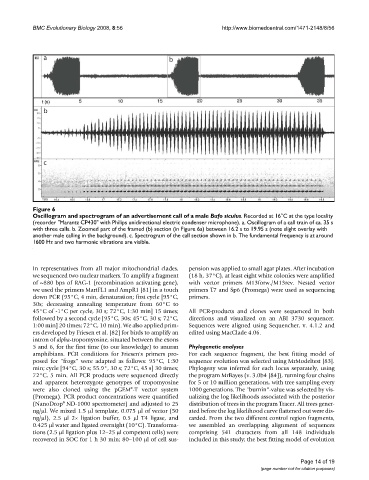
Figure 6
Oscillogram and spectrogram of an advertisement call of a male Bufo siculus
Oscillogram and spectrogram of an advertisement call of a male Bufo siculus. Recorded at 16°C at the type locality
(recorder "Marantz CP430" with Philips unidirectional electric condenser microphone). a. Oscillogram of a call train of ca. 35 s
with three calls. b. Zoomed part of the framed (b) section (in Figure 6a) between 16.2 s to 19.95 s (note slight overlay with
another male calling in the background). c. Spectrogram of the call section shown in b. The fundamental frequency is at around
1600 Hz and two harmonic vibrations are visible.
In representatives from all major mitochondrial clades, pension was applied to small agar plates. After incubation
we sequenced two nuclear markers. To amplify a fragment (18 h, 37°C), at least eight white colonies were amplified
of ~880 bps of RAG-1 (recombination activating gene), with vector primers M13forw./M13rev. Nested vector
we used the primers MartFL1 and AmpR1 [81] in a touch primers T7 and Sp6 (Promega) were used as sequencing
down PCR (95°C, 4 min, denaturation; first cycle [95°C, primers.
30s; decreasing annealing temperature from 60°C to
45°C of -1°C per cycle, 30 s; 72°C, 1:30 min] 15 times; All PCR-products and clones were sequenced in both
followed by a second cycle [95°C, 30s; 45°C, 30 s; 72°C, directions and visualized on an ABI 3730 sequencer.
1:00 min] 20 times; 72°C, 10 min). We also applied prim- Sequences were aligned using Sequencher, v. 4.1.2 and
ers developed by Friesen et al. [82] for birds to amplify an edited using MacClade 4.06.
intron of alpha-tropomyosine, situated between the exons
5 and 6, for the first time (to our knowledge) to anuran Phylogenetic analyses
amphibians. PCR conditions for Friesen's primers pro- For each sequence fragment, the best fitting model of
posed for "frogs" were adapted as follows: 95°C, 1:30 sequence evolution was selected using MrModeltest [83].
min; cycle [94°C, 30 s; 55.9°, 30 s; 72°C, 45 s] 30 times; Phylogeny was inferred for each locus separately, using
72°C, 5 min. All PCR products were sequenced directly the program MrBayes (v. 3.0b4 [84]), running four chains
and apparent heterozygote genotypes of tropomyosine for 5 or 10 million generations, with tree sampling every
®
were also cloned using the pGEM -T vector system 1000 generations. The "burnin"-value was selected by vis-
(Promega). PCR product concentrations were quantified ualizing the log likelihoods associated with the posterior
®
(NanoDrop ND-1000 spectrometer) and adjusted to 25 distribution of trees in the program Tracer. All trees gener-
ng/µl. We mixed 1.5 µl template, 0.075 µl of vector (50 ated before the log likelihood curve flattened out were dis-
ng/µl), 2.5 µl 2× ligation buffer, 0.5 µl T4 ligase, and carded. From the two different control region fragments,
0.425 µl water and ligated overnight (10°C). Transforma- we assembled an overlapping alignment of sequences
tions (2.5 µl ligation plus 12–25 µl competent cells) were comprising 541 characters from all 148 individuals
recovered in SOC for 1 h 30 min; 80–100 µl of cell sus- included in this study; the best fitting model of evolution
Page 14 of 19
(page number not for citation purposes)