Page 3 - Chrysoula_al_2010
P. 3
Historical dispersal of white sharks C. Gubili et al. 1681
GW44
GW10
GW43
GW8 GW9
GW40 GW39
GW37 7 GW35
GW15
GW12
2 2 4
2 GW42 43 GW14 GW11 GW6
GW7
GW41 GW36 GW17 GW2
GW38
GW13 GW1
GW4 GW3
missing haplotype
17 GW16
GW18
NW Atlantic GW32 GW33
Mediterranean GW31
GW45
GW30
South Africa 3 GW29 GW27
3
New Zealand 2
GW28
Australia GW34
GW24
NE Pacific GW25
GW23
2
GW22
GW20
GW26
GW19 2
GW21
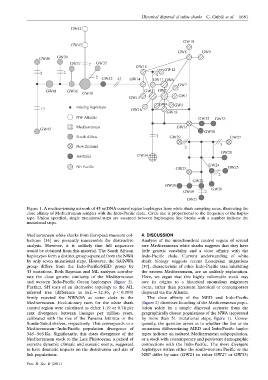
Figure 1. A median-joining network of 45 mtDNA control region haplotypes from white shark sampling areas, illustrating the
close affinity of Mediterranean samples with the Indo-Pacific clade. Circle size is proportional to the frequency of the haplo-
type. Unless specified, single mutational steps are assumed between haplotypes; line breaks with a number indicate the
mutational steps.
Mediterranean white sharks from European museum col- 4. DISCUSSION
lections [14] are presently inaccessible for destructive Analysis of the mitochondrial control region of several
analysis. However, it is unlikely that full sequences rare Mediterranean white sharks suggests that they have
would be obtained from this material. The South African little genetic variability and a close affinity with the
haplotypes form a distinct group separated from the NWA Indo-Pacific clade. Current understanding of white
by only seven mutational steps. However, the SA/NWA shark biology suggests recent Lessepsian migrations
group differs from the Indo-Pacific/MED group by [37], characteristic of other Indo-Pacific taxa inhabiting
43 mutations. Both Bayesian and ML analyses corrobo- the eastern Mediterranean, are an unlikely explanation.
rate the close genetic similarity of the Mediterranean Here, we argue that this highly vulnerable stock may
and western Indo-Pacific Ocean haplotypes (figure 2). owe its origins to a historical anomalous migratory
Further, SH tests of an alternative topology to the ML event, rather than persistent historical or contemporary
inferred tree (difference in ln L ¼ 32.36, p , 0.003) dispersal via the Atlantic.
firmly rejected the NWA/SA as sister clade to the The close affinity of the MED and Indo-Pacific
Mediterranean. Evolutionary rates for the white shark (figure 2) dismisses founding of the Mediterranean popu-
control region were calculated as either 1.19 or 0.74 per lation solely by a simple dispersal scenario from the
cent divergence between lineages per million years, geographically closest populations of the NWA (separated
calibrated with the rise of the Panama Isthmus or the by more than 51 mutational steps; figure 1). Conse-
Sunda-Sahul shelves, respectively. This corresponds to a quently, the question arises as to whether the five or six
Mediterranean–Indo-Pacific population divergence of mutations differentiating MED and Indo-Pacific haplo-
348–565 Ka. Significantly, this dates divergence of the types indicate an isolated Mediterranean subpopulation,
Mediterranean stock to the Late Pleistocene, a period of or a stock with contemporary and persistent demographic
extreme dynamic climatic and eustatic events, suggested connections with the Indo-Pacific. The most divergent
to have dramatic impacts on the distribution and size of haplotypes within either the south-western Pacific or the
fish populations. NEP differ by nine (GW21 to either GW27 or GW33)
Proc. R. Soc. B (2011)