Page 2 - Mullus1991
P. 2
720 M. CAMMARATA et al.
Tablc l. Allclic frequencics relative to thc analyzed loci and their R, values
Allclic frequencies
Tissue with M.b. M.s.
Enzymes Locus best aclivity R, No. 70 65
MDH MD Hl H, L, E, M, G 0.26 l
MDH2 L 0.33 l l
MDHJ H 0.18 o 0.138
LDH LDHI H, L, M, E 0.12 l l
LDH1 H, F, M 0.21 l l
LDH3 E 0.59 l l
ADH AD Hl L 0.12 0.028 o
ADH1 L 0.14 0.972 l
ADH2 L 0.26 0.028 o
ADH3 L 0.16 0.028 o
ADH4 L 0.19 l l
ADHS L 0.33 l l
ADH6 L 0.35 0.286 o
GPD GPD1 L 0.14 l l
GPD1 L, H, E 0.19 l
SOD SODI L 0.29 l l
SOD1 H,L,G 0.41 o l
SOD2 H,L,G 0.53 l o
IDH IDHl L 0.18 l l
IDH1 L 0.27 1 l
SDH SDHl L 0.28 1 l
SDH2 L 0.3 l l
l- 0.934 D =0.068
Generai protein
PMM PMMI M 0.31 l
PMM2 M 0.47 0.128
PMM3 M 0.61 l
Ali the above enzyme stammg solutions were then Genera/ proteins
incubated with gel slabs at 37"C unti! the appropriate bands
were visualized; staìnìng of the gels was stopped by rinsìng Analysis of generai proteins from liver, heart and
with water, and adding preservative solution (7% acetic eye has revealed such a high variance in the indi-
acid). Ali the products were purchased from Sigma. viduai patterns as to not be considered taxonomically
significant.
Generai protein analysis results far the muscle are
RESULTS characterized by the presence of the A and C (Fig. 2)
Isoenzymatic analysis areas which were abundant in labile bands and differ-
The seven enzymatic systems investigated for the
two species codify for the following 20 loci: MDH!,
MDH2, MDH3, LDHI, LDH2, LDH3, ADHI, LDH SOD
ADH2, ADH3, ADH4, ADH5, ADH6, ct.GPD l , a b
ct.GPD2, SOD l, SOD2, !D Hl, IDH2, SDHl,
SDH2. 1~ w
The allelic frequencies relative to the analyzed loci 2~
and their Rr are shown in Table l. There is increased
enzymatic activity in the organs considered.
In ali samples, l4loci appeared to be monomorphic ~
far the same allele (i.e. MDHl, MDH2, LDHI,
LDH2, LDH3, ADH4, ADH5, ct.GPDl, ct.GPD2, +-1
SOD l, JDH!, JDH2, SDHI, SDH2) and had
identica! patterns in the two species under study. }
The presence of MDH3 was limited ton. 9 (13.8%)
of Mullus surmuletus individuals, whereas in Mullus
barbatus ADHI, ADH3 and ADH6 were found in n. 2 : l' -
(2.8%), n. 2 (2.8%), and n. 20 (28.6%) of the speci- 3~ ·.
mens, respectively. In n. 2 (2.8%) of Mullus barbatus, ;.
the ADH2 locus presented an allelic variation.
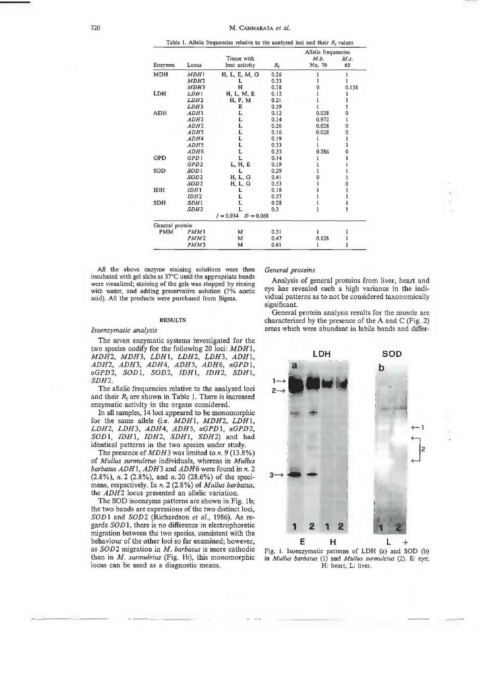
The SOD isoenzyme pattems are shown in Fig. l b;
the two bands are expressions of the two distinct loci,
SOD l and SOD2 (Richardson et al., 1986). As re-
gards SO D l, there is no difference in electrophoretic 1 2 1 2 :
migration between the two species, consistent with the
behaviour ofthe other Joci so far examined; however, E H l +
as SOD2 migration in M. barbatus is more cathodic Fig. l. Isoenzymatic patterns of LDH (a) and SOD (b)
than in M. surmuletus (Fig. lb), this monomorphic in Mullus barbatus (l) and Mullus surmuletus (2). E: eye;
locus can be used as a diagnostic means. H: heart; L: liver.