Page 6 - DNAfromhistorical
P. 6
238 Endang Species Res 27: 233–241, 2015
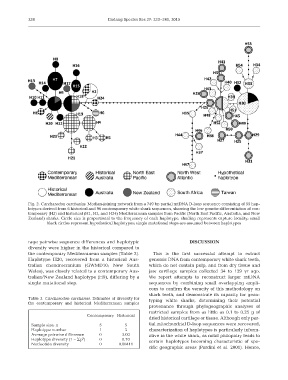
Fig. 2. Carcharodon carcharias. Median-joining network from a 749 bp partial mtDNA D-loop sequence consisting of 55 hap-
lotypes derived from 6 historical and 96 contemporary white shark sequences, showing the low genetic differentiation of con-
temporary (H2) and historical (H1, H2, and H24) Mediterranean samples from Pacific (North East Pacific, Australia, and New
Zealand) sharks. Circle size is proportional to the frequency of each haplotype; shading represents capture locality; small
black circles represent hypothetical haplotypes; single mutational steps are assumed between haplotypes
rage pairwise sequence differences and haplotypic DISCUSSION
diversity were higher in the historical compared to
the contemporary Mediterranean samples (Table 3). This is the first successful attempt to extract
Haplotype H20, recovered from a historical Aus- genomic DNA from contemporary white shark teeth,
tralian chondrocranium (GWMD10; New South which do not contain pulp, and from dry tissue and
Wales), was closely related to a contemporary Aus- jaw cartilage samples collected 34 to 129 yr ago.
tralian/New Zealand haplotype (H9), differing by a We report attempts to reconstruct larger mtDNA
single mutational step. sequences by combining small overlapping ampli-
cons to confirm the veracity of this methodology on
Table 3. Carcharodon carcharias. Estimates of diversity for shark teeth, and demonstrate its capacity for geno-
the contemporary and historical Mediterranean samples typing white sharks, determining their potential
provenance through phylogeographic analyses of
Contemporary Historical restricted samples from as little as 0.1 to 0.25 g of
dried historical cartilage or tissue. Although only par-
Sample size, n 5 5 tial mitochondrial D-loop sequences were recovered,
Haplotype number 1 3 characterization of haplotypes is particularly inform-
Average pairwise difference 0 3.00 ative in the white shark, as natal philopatry leads to
Haplotype diversity (1 − ∑p2) 0 0.70 certain haplotypes becoming characteristic of spe-
Nucleotide diversity 0 0.00411 cific geographic areas (Pardini et al. 2001). Hence,