Page 9 - Korn_alii_2006
P. 9
M. Korn et al. • Sister species within Triops cancriformis
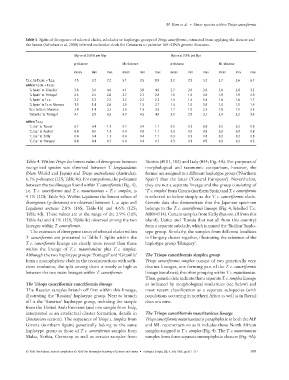
Table 5 Splits of divergence of selected clades, subclades or haplotype groups of Triops cancriformis, estimated from applying the slowest and
the fastest (Schubart et al. 2000) inferred molecular clock for Crustacea to pairwise 16S rDNA genetic distances.
Myr BP at 0.65% per Myr Myr BP at 0.9% per Myr
p-distance ML-distance p-distance ML-distance
mean min max mean min max mean min max mean min max
T.c.c. to T.c.m. + T.c.s. 4.5 3.2 7.2 5.1 3.5 8.9 3.2 2.3 5.2 3.7 2.6 6.4
within T.c.m. + T.c.s.:
‘S. Spain’ to ‘Gitanilla’ 3.8 3.6 4.0 4.1 3.9 4.5 2.7 2.6 2.9 3.0 2.8 3.2
‘S. Spain’ to ‘Portugal’ 2.5 2.5 2.6 2.7 2.7 2.8 1.8 1.8 1.9 1.9 1.9 2.0
‘S. Spain’ to T.c.s. 2.2 2.2 2.2 2.2 2.2 2.3 1.6 1.6 1.6 1.6 1.6 1.7
‘S. Spain’ to T.c.m. Morocco 1.9 1.4 2.6 2.0 1.5 2.7 1.4 1.0 1.9 1.4 1.1 1.9
T.c.s. to T.c.m. Morocco 2.4 1.4 3.2 2.5 1.5 3.5 1.7 1.0 2.3 1.8 1.1 2.5
‘Gitanilla’ to ‘Portugal’ 4.1 3.9 4.3 4.7 4.5 4.9 3.0 2.9 3.1 3.4 3.2 3.5
within T.c.c.:
‘C. Eur.’ to ‘Russia’ 0.7 0.4 1.1 0.7 0.4 1.1 0.5 0.3 0.8 0.5 0.3 0.8
‘C. Eur.’ to ‘Austria’ 0.4 0.0 1.1 0.4 0.0 1.1 0.3 0.0 0.8 0.3 0.0 0.8
‘C. Eur.’ to ‘Sicily’ 0.4 0.4 1.1 0.4 0.4 1.1 0.3 0.3 0.8 0.3 0.3 0.8
‘C. Eur.’ to ‘Hungary’ 0.4 0.4 0.7 0.4 0.4 0.7 0.3 0.3 0.5 0.3 0.3 0.5
Table 4. Within Triops the lowest value of divergence between Tunisia (#101, 102) and Italy (#34; Fig. 4A). For purposes of
recognized species was observed between T. longicaudatus morphological and taxonomic comparison, however, the
(New World and Japan) and Triops australiensis (Australia): former are assigned to a different haplotype group (‘Northern
6.1% p-distance (12S; Table 4c). For comparison, the p-distance Spain’) than the latter (‘Central European’). Nevertheless,
between the two lineages found within T. cancriformis (Fig. 4), they are not a separate lineage and the group consisting of
i.e. T. c. cancriformis and T. c. mauritanicus + T. c. simplex, is ‘T. c. simplex’ from Girona (northern Spain) and T. c. cancriformis
4.1% (12S; Table 4c). Within Lepidurus the lowest values of is referred to below simply as the T. c. cancriformis clade.
divergence (p-distance) are observed between L. a. apus and Genetic data also demonstrate that the Japanese specimen
Lepidurus arcticus: 2.8% (16S; Table 4b) and 4.6% (12S; belongs to the T. c. cancriformis lineage (Fig. 4, labelled T.c.
Table 4d). These values are in the range of the 2.9% (16S; AB084514). Certain samples from Sicily (but not all from this
Table 4a) and 4.1% (12S; Table 4c) observed among the two island), Ustica and Tunisia (but not all from this country)
lineages within T. cancriformis. form a separate subclade, which is named the ‘Sicilian’ haplo-
The estimates of divergence times of selected clades within type group. Similarly, the samples from different localities
T. cancriformis are presented in Table 5. Splits within the in Hungary cluster together, illustrating the existence of the
T. c. cancriformis lineage are clearly more recent than those haplotype group ‘Hungary’.
within the lineage of T. c. mauritanicus plus T. c. simplex.
Although the two haplotype groups ‘Portugal’ and ‘Gitanilla’ The Triops cancriformis simplex group
form a monophyletic clade in the reconstructions with suffi- Triops cancriformis simplex consist of two genetically very
cient resolution, the split among them is nearly as high as distinct lineages, one forming part of the T. c. cancriformis
between the two main lineages within T. cancriformis. lineage (see above), the other grouping within T. c. mauritanicus.
Thus, genetic data indicate that a separate T. c. simplex lineage
The Triops cancriformis cancriformis lineage as indicated by morphological similarities (see below) and
The Russian samples branch off first within this lineage, most recent classification as a separate subspecies (with
illustrating the ‘Russian’ haplotype group. Next to branch populations occurring in northern Africa as well as in Iberia)
off is the ‘Austrian’ haplotype group, including the sample does not exist.
from the United Arab Emirates (and one sample from Italy,
interpreted as an artefactual cluster formation, details in The Triops cancriformis mauritanicus lineage
Discussion section). The sequences of Triops c. simplex from Triops cancriformis mauritanicus is paraphyletic in both the MP
Girona (northern Spain) genetically belong to the same and ML reconstructions as it includes those North African
haplotype group as those of T. c. cancriformis samples from samples assigned to T. c. simplex (Fig. 4). The T. c. mauritanicus
Malta, Serbia, Germany as well as certain samples from samples form three separate monophyletic clusters (Fig. 4A):
© 2006 The Authors. Journal compilation © 2006 The Norwegian Academy of Science and Letters • Zoologica Scripta, 35, 4, July 2006, pp301–322 309