Page 6 - Korn_alii_2006
P. 6
Sister species within Triops cancriformis • M. Korn et al.
analysis, where haplotype groups are defined as subgroups of
sequences sharing autapomorphic sites, without considera-
tion of singletons (Appendix 1). There was one exception
from grouping specimens by haplotype group: the northern
Spanish population was treated like a separate haplotype group
to evaluate its unusual position in the present classification in
which it is assigned to a subspecies (i.e. T. c. simplex) different
to the other members of its haplotype group (T. c. cancriformis).
The dependent variables were telson length ratio, number of
apodous abdominal segments and number of dorsal carina
spines, respectively. To test for homogeneity of variance,
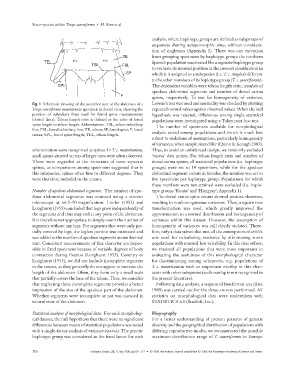
Fig. 1 Schematic drawing of the posterior part of the abdomen of a Levene’s test was used and normality was checked by plotting
Triops cancriformis mauritanicus specimen in dorsal view, showing the expected normal values against observed values. When the null
position of subsidiary lines used for furcal spine measurements hypothesis was rejected, differences among single statistical
(dotted lines). Telson length ratio is defined as the ratio of furcal populations were investigated using a Tukey post-hoc test.
spine length to telson length. Abbreviations: TSL, telson subsidiary The number of specimens available for morphological
line; FSL, furcal subsidiary line; TE, telson; SP, furcal spine; F, furcal
analysis varied among populations and ANOVA is much less
ramus; SPL, furcal spine length; TEL, telson length.
robust to violations of assumptions, particularly homogeneity
of variances, when sample sizes differ (Quinn & Keough 2003).
sclerotization were recognized as spines. In T. c. mauritanicus, Thus, to avoid an unbalanced design, we randomly excluded
small spines situated on top of larger ones were often observed. ‘excess’ data points. For telson length ratio and number of
These were regarded as the remnants of once separate dorsal carina spines, all statistical populations (i.e. haplotype
spines, as comparisons among specimens suggested that in groups) were set to 10 specimens, while for the apodous
this subspecies, spines often fuse to different degrees. They abdominal segment counts in females, the number was set to
were therefore included in the counts. five specimens per haplotype group. Populations for which
these numbers were not attained were excluded (i.e. haplo-
Number of apodous abdominal segments. The number of apo- type groups ‘Russia’ and ‘Hungary’; Appendix 1).
dous abdominal segments was counted using a stereo- The dorsal carina spine counts showed positive skewness,
microscope at ×6.5–50 magnification. Linder (1952) and resulting in nonhomogeneous variances. Thus, a square root
Longhurst (1955) concluded that legs grow independently of transformation was used, which greatly improved the
the segments and thus may end at any point of the abdomen. approximation to a normal distribution and homogeneity of
It is therefore not appropriate to simply count the number of variances within this dataset. However, the assumption of
segments without any legs. For segments that were only par- homogeneity of variances was still clearly violated. There-
tially covered by legs, the legless portion was estimated and fore, only a data subset that met all the assumptions of ANOVA
was added to the number of apodous segments as one decimal was used for calculating statistics by eliminating some
unit. Consistent measurements of this character are impos- populations with unusual low variability. In this data subset,
sible in fixed specimens because of variable degrees of body we retained all populations that were most important in
contraction during fixation (Longhurst 1955). Contrary to evaluating the usefulness of this morphological character
Longhurst (1955), we did not include incomplete segments for discriminating among subspecies, e.g. populations of
in the counts, as they generally do not appear to increase the T. c. mauritanicus with an important overlap in this char-
length of the abdomen. Often, they form only a small scale acter with other subspecies (such overlap is not recognized in
that partially covers the base of the telson. Thus, we consider the present literature).
that neglecting these incomplete segments provides a better Following data analysis, a sequential Bonferroni test (Rice
impression of the size of the apodous part of the abdomen. 1989) was carried out for the three ANOVAs performed. All
Whether segments were incomplete or not was assessed in statistics on morphological data were undertaken with
ventral view of the abdomen. STATISTICA 6.0 (StatSoft, Inc.).
Statistical analyses of morphological data. For each morpholog- Biogeography
ical dataset, the null hypothesis that there were no significant For a better understanding of present patterns of genetic
differences between means of statistical populations was tested diversity and the geographical distribution of populations with
with a single-factor analysis of variance (ANOVA). The genetic differing reproductive modes, we reconstructed the possible
haplotype group was considered as the fixed factor for each maximum distribution range of T. cancriformis in Europe
306 Zoologica Scripta, 35, 4, July 2006, pp301–322 • © 2006 The Authors. Journal compilation © 2006 The Norwegian Academy of Science and Letters