Page 5 - Passalacqua_Peruzzi_Pellegrino2008
P. 5
TAXON 57 (3) • August 2008: 893–906 Passalacqua & al. • Biosystematics of the Jacobaea maritima group
(Norušis, 2005). Multivariate analysis was performed using
Gower coefficient for mixed data extended to ordinal char-
acters (Podani, 1999). Continuous characters were subject
to analysis of variance (ANOVA) after Bloom’s normalisa-
tion, and F statistics was used to select significant charac-
ters (P < 0.001). The Kruskal Wallis test (H ) was used for
continuous characters that did not respond to normality or
homoscedasticy tests, and for semiquantitative characters.
Binary characters were subject to a Pearson chi square sta-
2
tistic test ( χ ). Pairwise deletion was used for missing data.
All statistical values are given in Appendix 2.
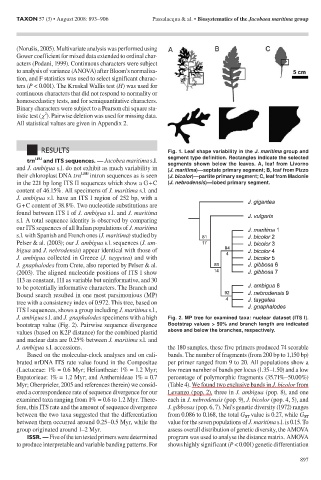
RESULTS Fig. 1. Leaf shape variability in the J. maritima group and
trn LEU and ITS sequences. — Jacobea maritima s.l. segment type definition. Rectangles indicate the selected
LEU
and J. ambigua s.l. do not exhibit as much variability in segments shown below the leaves. A, leaf from Livorno
(J. maritima)—septate primary segment; B, leaf from Pizzo
LEU
their chloroplast DNA trn LEU intron sequences as is seen (J. bicolor)—partite primary segment; C, leaf from Madonie
in the 221 bp long ITS II sequences which show a G + C (J. nebrodensis)—lobed primary segment.
content of 46.15%. All specimens of J. maritima s.l. and
J. ambigua s.l. have an ITS I region of 252 bp, with a
G + C content of 38.8%. Two nucleotide substitutions are
found between ITS I of J. ambigua s.l. and J. maritima
s.l. A total sequence identity is observed by comparing
our ITS sequences of all Italian populations of J. maritima
s.l. with Spanish and French ones (J. maritima) studied by
Pelser & al. (2003); our J. ambigua s.l. sequences (J. am-
bigua and J. nebrodensis) appear identical with those of
J. ambigua collected in Greece (J. taygetea) and with
J. gnaphalodes from Crete, also reported by Pelser & al.
(2003). The aligned nucleotide positions of ITS I show
113 as constant, 111 as variable but uninformative, and 30
to be potentially informative characters. The Branch and
Bound search resulted in one most parsimonious (MP)
tree with a consistency index of 0.972. This tree, based on
ITS I sequences, shows a group including J. maritima s.l.,
J. ambigua s.l. and J. gnaphalodes specimens with a high Fig. 2. MP tree for examined taxa: nuclear dataset (ITS I).
bootstrap value (Fig. 2). Pairwise sequence divergence Bootstrap values > 50% and branch length are indicated
values (based on K2P distance) for the combined plastid above and below the branches, respectively.
and nuclear data are 0.25% between J. maritima s.l. and
J. ambigua s.l. accessions. the 180 samples, these five primers produced 74 scorable
Based on the molecular-clock analyses and on cali- bands. The number of fragments (from 200 bp to 1,150 bp)
brated nrDNA ITS rate value found in the Compositae per primer ranged from 9 to 20. All populations show a
(Lactuceae: 1% = 0.6 Myr; Heliantheae: 1% = 1.2 Myr; low mean number of bands per locus (1.35–1.50) and a low
Eupatorieae: 1% = 1.2 Myr; and Anthemideae 1% = 0.7 percentage of polymorphic fragments (35.71%–50.00%)
Myr; Oberprieler, 2005 and references therein) we consid- (Table 4). We found two exclusive bands in J. bicolor from
ered a correspondence rate of sequence divergence for our Levanzo (pop. 2), three in J. ambigua (pop. 8), and one
examined taxa ranging from 1% = 0.6 to 1.2 Myr. There- each in J. nebrodensis (pop. 9), J. bicolor (pop. 4, 5), and
fore, this ITS rate and the amount of sequence divergence J. gibbosus (pop. 6, 7). Nei’s genetic diversity (1972) ranges
between the two taxa suggested that the differentiation from 0.086 to 0.168, the total G STST value is 0.27, while G STST
between them occurred around 0.25–0.5 Myr, while the value for the seven populations of J. maritima s.l. is 0.15. To
group originated around 1–2 Myr. assess overall distribution of genetic diversity, the AMOVA
ISSR. — Five of the ten tested primers were determined program was used to analyse the distance matrix. AMOVA
to produce interpretable and variable banding patterns. For shows highly significant (P < 0.001) genetic differentiation
897