Page 4 - Colliard_ali_2010
P. 4
Colliard et al. BMC Evolutionary Biology 2010, 10:232 Page 4 of 16
http://www.biomedcentral.com/1471-2148/10/232
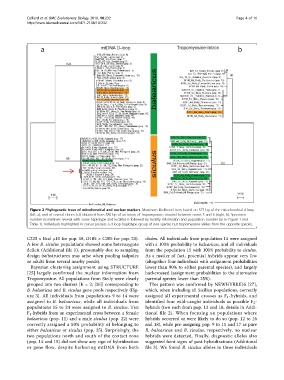
Figure 2 Phylogenetic trees of mitochondrial and nuclear markers. Maximum likelihood trees based on 577 bp of the mitochondrial d-loop
(left, a), and of several clones (cl.) obtained from 580 bp of an intron of tropomyosine, situated between exons 5 and 6 (right, b). Specimen
number (sometimes several with same haplotype and locality) is followed by locality information and population number (as in Figure 1 and
Table 1). Individuals highlighted in colour possess a d-loop haplotype group of one species but tropomyosine alleles from the opposite species.
C223 × Bcal μ10 for pop. 18, D105 × C205 for pop. 23). clades. All individuals from population 14 were assigned
Afew B. siculus populations showed some heterozygote with a 100% probability to balearicus, and all individuals
deficit (Additional file 1), presumably due to sampling from the population 15 with 100% probability to siculus.
design (substructures may arise when pooling tadpoles As a matter of fact, potential hybrids appear very few
or adults from several nearby ponds). (altogether four individuals with assignment probabilities
Bayesian clustering assignment using STRUCTURE lower than 90% to either parental species), and largely
[25] largely confirmed the nuclear information from backcrossed (assignment probabilities to the alternative
Tropomyosine. All populations from Sicily were clearly parental species lower than 25%).
grouped into two clusters [K = 2; [26]] corresponding to This pattern was confirmed by NEWHYBRIDS [27],
B. balearicus and B. siculus gene pools respectively (Fig- which, when including all Sicilian populations, correctly
ure 3). All individuals from populations 9 to 14 were assigned all experimental crosses as F 1 -hybrids, and
assigned to B. balearicus, while all individuals from identified four wild-caught individuals as possible F 2 -
populations 15 to 24 were assigned to B. siculus.Ten hybrids (two each from pop. 13 and 18, details in Addi-
F 1 -hybrids from an experimental cross between a female tional file 2). When focusing on populations where
balearicus (pop. 11) and a male siculus (pop. 22) were hybrids occurred or were likely to do so (pop. 12 to 16
correctly assigned a 50% probability of belonging to and 18), while pre-assigning pop. 9 to 11 and 17 as pure
either balearicus or siculus (pop. 25). Surprisingly, the B. balearicus and B. siculus, respectively, no nuclear
two populations north and south of the contact zone hybrids were detected. Finally, diagnostic alleles also
(pop. 14 and 15) did not show any sign of hybridization suggested faint signs of past hybridizations (Additional
or gene flow, despite harboring mtDNA from both file 3). We found B. siculus alleles in three individuals